

The INDRA Biomedical Discovery Engine is built on INDRA CoGEx, a graph database integrating causal relations, ontological relations, properties, and data, assembled at scale automatically from the scientific literature and structured sources.
Biological Entities
Clinical Trials
Publications
Evidences
Research Projects
Patents
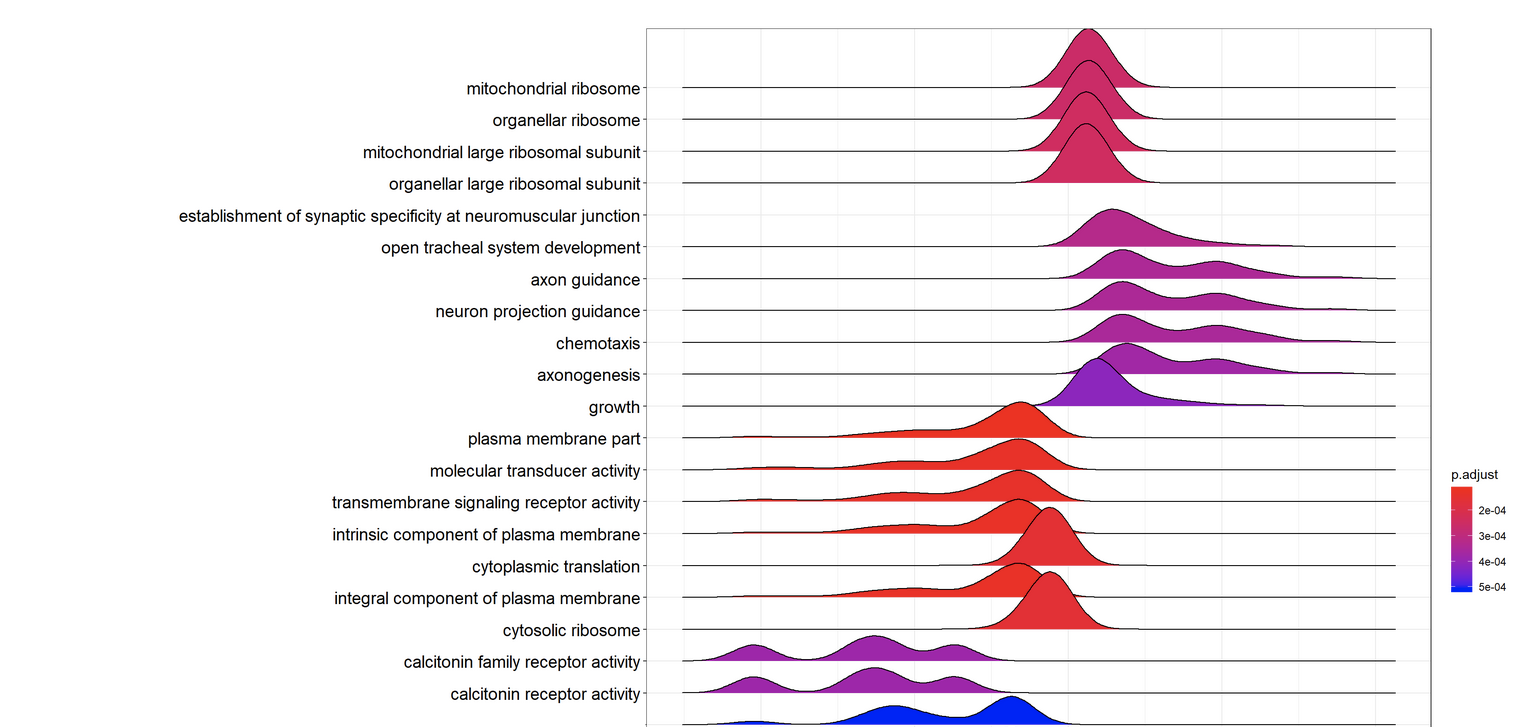
Performs gene set enrichment analysis using INDRA CoGEx.
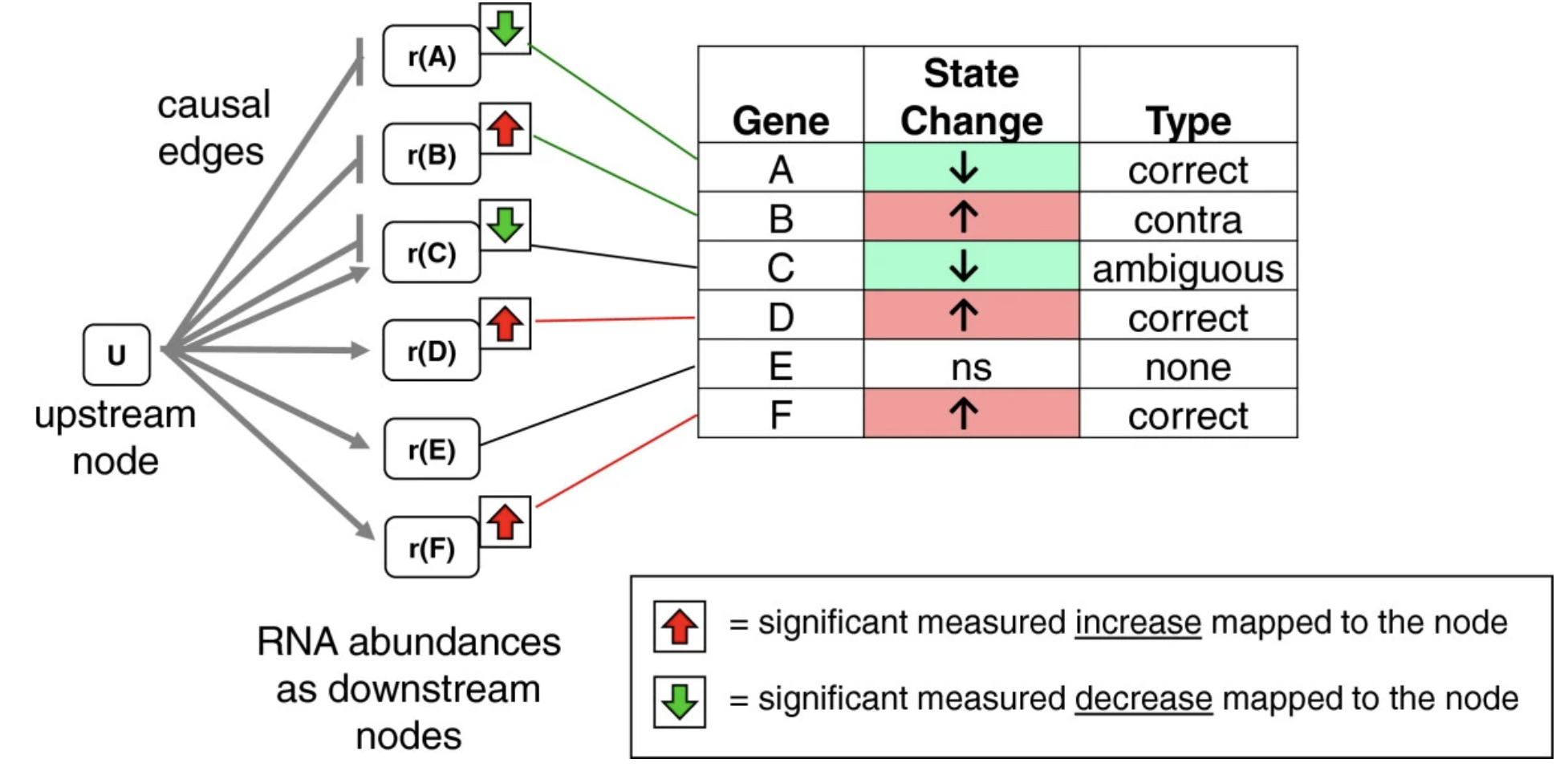
Perform signed gene set enrichment analysis using INDRA CoGEx and the Reverse Causal Reasoning algorithm.
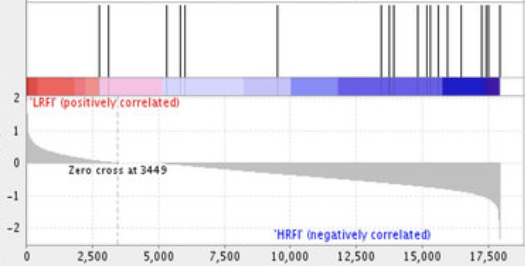
Perform gene set enrichment analysis on continuous data using INDRA CoGEx.
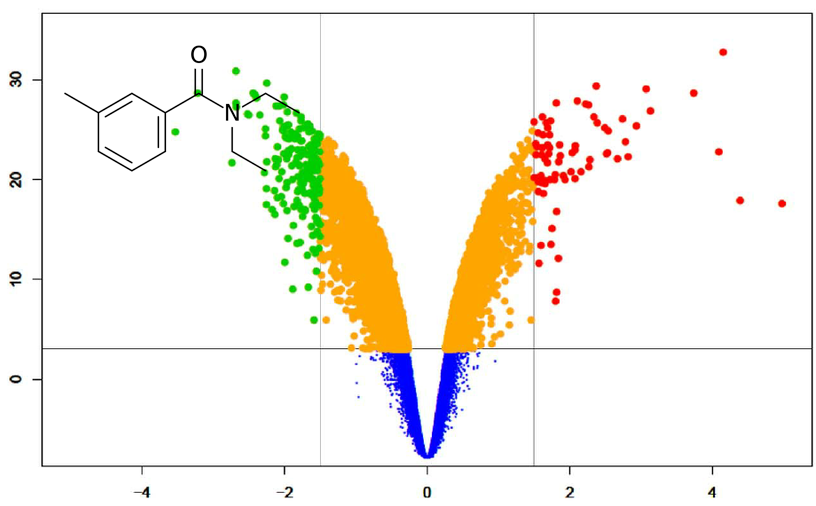
Perform metabolite set enrichment analysis using INDRA CoGEx.
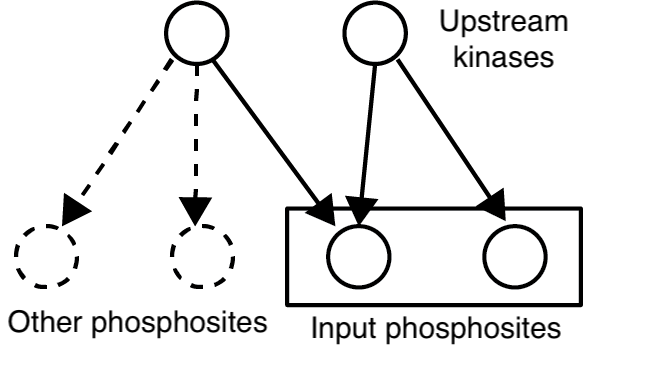
Identify kinases that are statistically enriched in a set of phosphosites based on known kinase-substrate relationships.
Analyze relationships between a source gene and its downstream target genes.
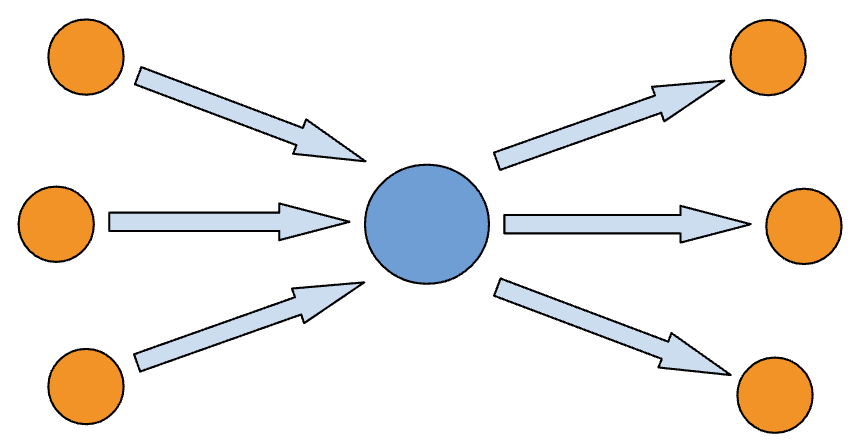
Search statements assembled from literature and databases by agents involved.
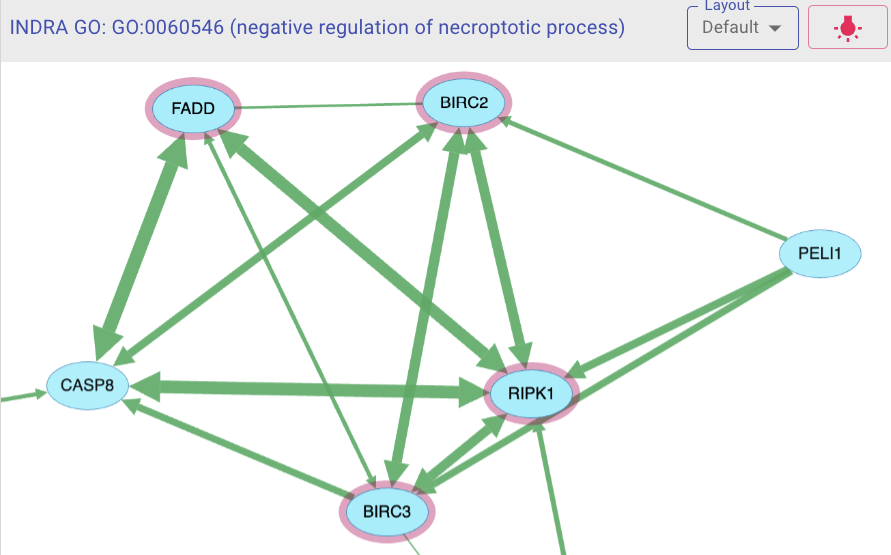
Explore networks induced by genes associated with a given Gene Ontology term.

Explore statements from publications annotated with the given MeSH term.

Explore statements from a given publication.

Explore statements for the subnetwork induced by given entities.
Explore statements about transcription factors regulating the amounts of their targets not already appearing in databases like Pathway Commons.

Explore statements about kinases phosphorylating targets not already appearing in databases like PhosphoSitePlus.

Explore statements about phosphatases dephosphorylating targets not already appearing in databases like Pathway Commons.
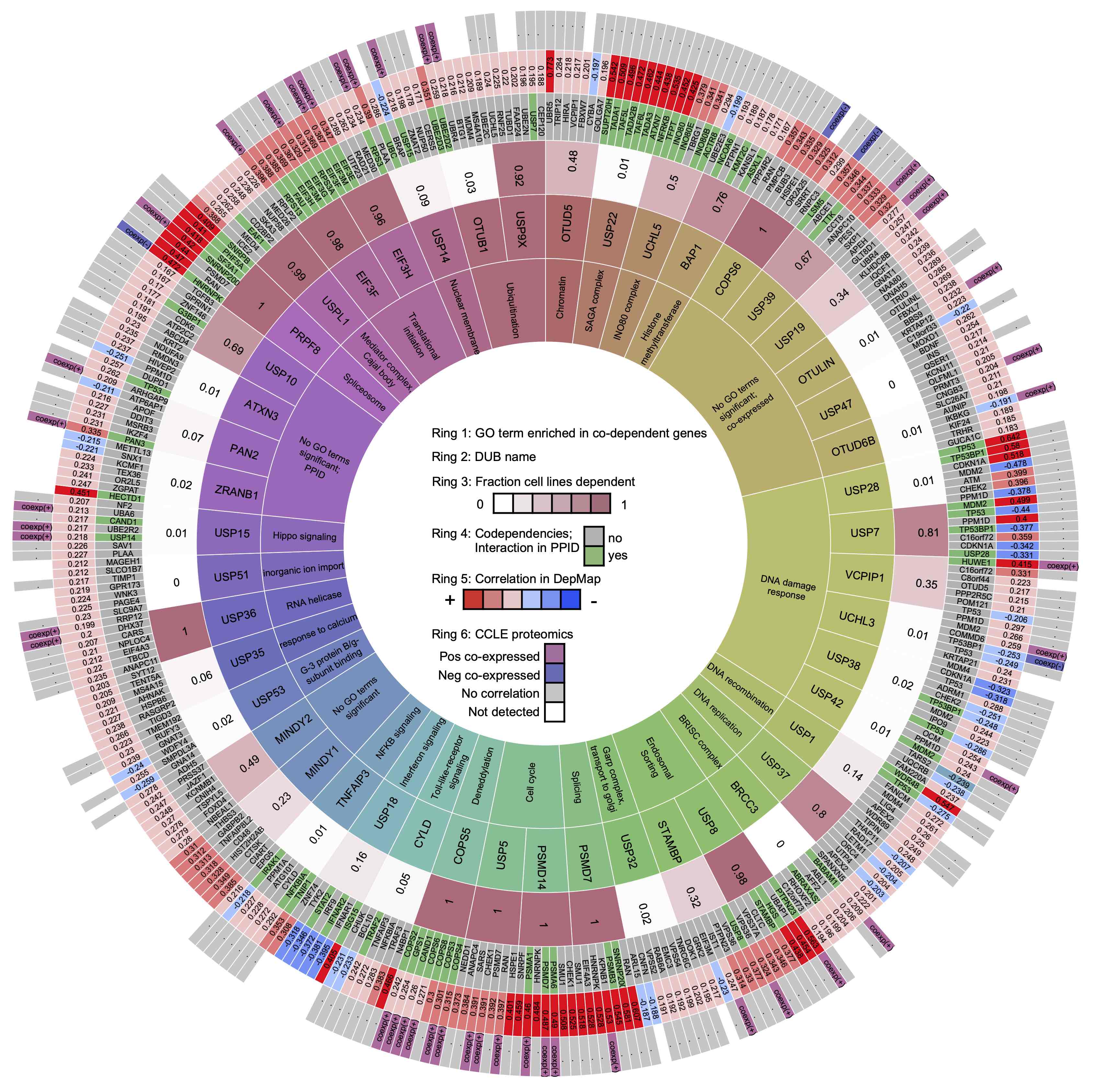
Explore statements about deubiquitinating enzymes.

Explore gene-GO term associations that are not already available as GO annotations.

Explore protein-protein interactions not already appearing in databases like BioGRID.

Explore miRNA-target interactions not already appearing in databases like miRTarBase.
Explore intrensically disordered proteins' interactions not already appearing in databases.